AUTS2-Related Neuropsychiatric Disorders with Intrafamilial Variability
Fiorella Gurrieri1*, Alessia Azzara2, Ilaria Cassano2, Elisa Paccagnella2, Carla Lintas2, Luigi Laino3, Paola Grammatico3 and Roberto Sacco4
1Department of Medical Genetics, Campus Bio-Medico University, Rome, Italy
2Department of Life Sciences and Public Health, University of the Sacred Heart, Rome, Italy
3Department of Medical Genetics, University of S Camillo-Forlanini Hospital, Rome, Italy
4Department of Child Neuropsychiatry, Campus Bio-Medico University, Rome, Italy
- *Corresponding Author:
- Fiorella Gurrieri, Department of Medical Genetics, Campus Bio-Medico University, Rome, Italy, Tel: 39906225419174; E-mail: a.azzara@unicampus.it
Received date: February 07, 2022, Manuscript No. IPJNP-22-12680; Editor assigned date: February 10, 2022, PreQC No. IPJNP-22-12680 (PQ); Reviewed date: February 25, 2022, QC No. IPJNP-22-12680; Revised date: April 11, 2022, Manuscript No. IPJNP-22-12680 (R); Published date: April 19, 2022, DOI: 10.36648/2471-8548.22.06.001
Citation: Gurrieri F, Azzara A, Cassano I, Paccagnella E, Lintas C, et al. (2022) AUTS2-Related Neuropsychiatric Disorders with Intrafamilial Variability. J Neuropsychiatry Vol:6 No:4
Abstract
Intellectual disability, autism and psychiatric conditions are a group of clinically and genetically heterogeneous neurodevelopmental disorders. Their pathogenesis involves many genes responsible for neuronal migration, axon extension, synaptic function and transcriptional regulation. Among them, autism susceptibility candidate 2 (AUTS2, OMIM *607270) gene has been recurrently associated with syndromic intellectual disability and autism: both single nucleotide and intragenic exonic deletions have been reported. Genotype-phenotype correlation studies suggest that C-terminal disruptions have a more severe phenotype with respect to N-terminal ones.
We report on a family segregating and intronic variant with functional effects in AUTS2, showing marked intrafamilial variability at the clinical level in three carrier members: the proposita, a 24-years-old girl with developmental delay, severe hyperactivity, sleep apnoea, support teacher from early on and self-harmful behaviours; her 20-years-old brother with hyperactivity, cognitive impairment and developmental motor delay; the mother, who displays peculiar facial features, strikingly resembling those reported in AUTS2 patients, and referred a support teacher at school.
Intronic deletions in AUTS2 segregating with the known phenotype are not acknowledged as all the reported deletions encompass exons: therefore, the deletion observed in our family might be crucial to identify new regulatory sequences. The patient’s deletion, even though it is located far from the intron-exon boundary, includes two CpG islands and one regulatory element, whose removal might impair the appropriate expression of AUTS2. Besides that, RNA studies confirmed an exon skipping resulting in the loss of eighteen aminoacid residues in the N- terminal region.
Keywords
AUTS2; Intronic deletions; Syndromic autism
Introduction
Neuro Developmental Disorders (NDD), including intellectual disability, autism and psychiatric disorders are a group of clinically and genetically heterogeneous conditions. Their pathogenesis involves many genes responsible for neuronal migration, axon extension, synaptic function and transcriptional regulation. An increasing number of such genes are being identified because of the application in the diagnostic routine of pangenomic tests, such as multigene panels or exam analysis as well as array-CGH. Among these genes, autism susceptibility candidate 2 (AUTS2, OMIM*607270), involved in neural migration and neurogenesis has been recurrently associated with syndromic intellectual disability but the phenotype is not easily recognizable yet [1,2]. Disruption of AUTS2 causes a syndrome form of Intellectual Disability (ID) known as “AUTS2 syndrome” (OMIM#615834), characterized by a highly variable phenotype consisting of Global Developmental Delay (GDD) commonly associated with the combination of microcephaly, short stature, feeding difficulties and hypotonic, as well as recognizable facial dysmorphic features [3]. In literature, it is observed large interindividual, and even intrafamilial, variability in patients with AUTS2 pathogenic variants. More than 60 patients have been described since the identification of the gene mostly carrying de novo deletions of one or more exons, while pathogenic single nucleotide variants only represent a small fraction of the mutational spectrum.
In this study, we report on the first exclusively intronic deletion of the AUTS2 gene segregating in 3 differently affected family members, whose clinical features range from just mild learning disability to severe cognitive impairment, psychiatric disturbance and characteristic facial findings. We also provide further evidence of interfamilial phenotypic variability in AUTS2 syndrome.
Case presentation
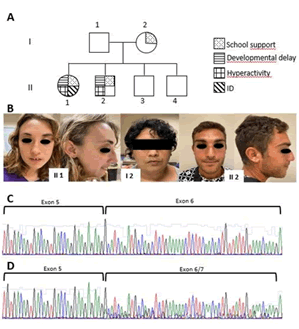
A 24-years-old girl born from non-consanguineous parents was referred because of significant intellectual disability and behavioural issues of unknown an etiology. After birth, no medical problems were reported. She did not start walking until eighteen months of age, had normal speech development with few words acquisition. She was diagnosed with developmental delay since the age of three with support teacher from nursery school. She suffered of sleep apnoea. Since she was sixteen, she developed psychiatric illness, self-harmful behaviour and hyperactivity. CT scan, EEG and ECG were normal. Her physical examination showed dysmorphic facial features with microcephaly (OFC 51.5 cm, -3SD), short stature (155 cm, 10°-25°centile), elongated face, short and upward eyelid rims, irregular eyebrows, proptosis, prominent nasal bridge with hypoplastic wings, short philtrum and mouth with thin mucous border, highly arched palate, irregular dentition, malar hypoplasia, normal ears; hands appeared with gnarled fingers and distal ends widened. In her family, a 20-years-old brother showed a milder but clinically noticeable clinical problem with mild hyperactivity, learning difficulties and developmental motor delay. They had two other younger healthy brothers. At our family evaluation, their mother referred she needed a support teacher at school and, upon clinical evaluation; she showed similar peculiar facial features (Figure 1 A, B).
Methods and Results
Genomic DNA was extracted from peripheral blood of the proposita, her affected brother and both parents, to perform array-CGH (Cynosures Kit, 8 × 60K). Array-CGH analysis revealed a 141 kb heterozygous copy number loss involving a part of intron 5 of the AUTS2 gene: arr (GRCh37) 7q11.22 (69974660-70115654) x 1 with proximal and distal breakpoints distant about 73 Kb and 48 Kb, respectively, from intron-exon boundary and classified as a Variant of Unknown Significance (VUS). The segregation analysis revealed that the micro deletion was maternally inherited and also present in the affected brother. Phenotype re-evaluation suggested that it strikingly resembled the one observed in AUTS2 patients, and therefore we performed cDNA sequence analysis which revealed that the two affected siblings and their mother showed a heterozygous skipping of exon 6 (Figure 1 C,D). In order to rule out that phenotypic variability in the affected family members could be due to additional intragenic variants, we sequenced the entire AUTS2 gene, but no variant was detected.
Discussion
In this study, through an array-CGH analysis, we identified an intrinsic deletion of the AUTS2 gene causing the skipping of exon 6 in AUTS2 transcript. Segregation analysis showed that the genomic micro deletion and the skipping of exon 6 were present in all affected individuals, while absent in the unaffected father.
AUTS2 is located on chromosome 7q11.22, a region associated with susceptibility to chromosomal breakpoints; it spans 1.9 Mb on the genomic DNA and consists of 19 exons. The gene may be divided in two parts: the first half of the gene (exons 1-6) has relatively large introns, the second half (C- terminus) has smaller clustered introns (exons 7-19) [4,5]. The pathogenic Copy Number Variations (CNVs) within AUTS2 are well distributed in either the fast-evolving 5’ end or within the highly conserved C-terminus region.
In AUTS2 syndrome two distinct phenotypes in correlation to genotype have been described, with C-terminal disruption causing more severe phenotype. Patients with 3’ deletions also display more pronounced dysmorphic features, while patients with 5’ in-frame deletions have a mild phenotype or may even be unaffected [6,7]. In-frame deletions have been reported in mildly or unaffected parents of index patients with 5’ in-frame deletions, whereas exon 6 deletions are out-of-frame and can cause severe phenotypes [8-10].
To date, ten patients with exon 6 deletions have been described all affected by variable intellectual disability/developmental delay. Intronic deletions in AUTS2 that segregate with the known phenotype are not known yet (all the reported ones in the literature encompass exons): therefore, the deletion observed in our family appears to be crucial to the phenotype and uncovers potential regulatory regions of this gene. This intronic deletion, that includes two CpG islands and one regulatory element, might impair the transcription with depletion of exon 6 and inappropriate expression of AUTS2.
In the literature, patients with involvement of exon 6 are all affected to a greater or lesser extent but no cases without cognitive involvement have been described. In our family, we have the simultaneous presence of a very mildly affected member, with only learning difficulties, and two markedly affected members with varying severity: this might be due to the influence of the individual genetic background or to some imprinting effect. Furthermore, deep intronic variants with proven exon skipping have never been described, therefore the deletion was considered as a VUS. Interestingly, the communication between the clinic and the laboratory and phenotype re-evaluation has raised the suspicion of the pathogenicity of this variant, and subsequently prompted the diagnostic deepening at transcription level, with reclassification of the variant in class 4 (probably pathogenetic variant) following its demonstrated impact on cDNA.
Conclusion
The possibility of analyzing patients with a poorly recognizable clinical picture by means of pangenomic tests will allow the identification of numerous variants of difficult interpretation. As demonstrated by this report, the cross-talk between the clinic and the laboratory is crucial to orient the VUS classification.
We here report a previously undescribed disease mechanism for AUTS2-related intellectual disability, i.e. a pathogenic deep intronic deletion, far from the splicing site: this suggest that in most instances phenotype analysis is mandatory to increase the diagnostic yield of pangenomic tests.
References
- Palumbo P, Di Muro E, Accadia M, Benvenuto M, Di Giacomo MC, et al. (2021) Whole exome sequencing reveals a novel AUTS2 in-frame deletion in a boy with global developmental delay, absent speech, dysmorphic features, and cerebral anomalies. Genes (Basel) 12:1–10
[Crossref] [Google Scholar] [Pubmed]
- Beunders G, Voorhoeve E, Golzio C, Pardo LM, Rosenfeld JA, et al. (2013) Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am J Hum Genet 92:210–220
[Crossref] [Google Scholar] [Pubmed]
- Sanchez-Jimeno C, Blanco-Kelly F, Lopez-Grondona F, Losada-Del Pozo R, Moreno B, et al. (2021) Attention deficit hyperactivity and autism spectrum disorders as the core symptoms of auts2 syndrome: Description of five new patients and update of the frequency of manifestations and genotype-phenotype correlation. Genes (Basel) 12:1–10
[Crossref] [Google Scholar] [Pubmed]
- Gieldon L, Jauch A, Obeid K, Kaufmann L, Hinderhofer K, et al. (2021) Germ cell mosaicism for AUTS2 exon 6 deletion. Am J Med Genet Part A 185:1261–1265
[Crossref] [Google Scholar] [Pubmed]
- Amarillo IE, Li WL, Li X, Vilain E, Kantarci S (2014) De novo single exon deletion of AUTS2 in a patient with speech and language disorder: A review of disrupted AUTS2 and further evidence for its role in neurodevelopmental disorders. Am J Med Genet Part A 164:958–965
[Crossref] [Google Scholar] [Pubmed]
- Oksenberg N, Ahituv N (2013) The role of AUTS2 in neurodevelopment and human evolution. Trends Genet 29:600–608
[Crossref] [Google Scholar] [Pubmed]
- Saeki S, Enokizono T, Imagawa K, Fukushima H, Kajikawa D, et al. (2019) A case of autism spectrum disorder with cleft lip and palate carrying a mutation in exon 8 of AUTS2. Clin Case Rep 7:2059–2063
[Crossref] [Google Scholar] [Pubmed]
- Beunders G, De Munnik SA, Van Der Aa N, Ceulemans B, Voorhoeve E, et al. (2015) Two male adults with pathogenic AUTS2 variants, including a two-base pair deletion, further delineate the AUTS2 syndrome. Eur J Hum Genet 23:803–807
[Crossref] [Google Scholar] [Pubmed]
- Asadollahi R, Oneda B, Joset P, Azzarello-Burri S, Bartholdi D, et al. (2014) The clinical significance of small copy number variants in neurodevelopmental disorders. J Med Genet 51:677–688
[Crossref] [Google Scholar] [Pubmed]
- Hori K, Hoshino M (2017) Neuronal migration and AUTS2 syndrome. Brain Sci 7:54
[Crossref] [Google Scholar] [Pubmed]

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences